The brain is composed of a wide array of interconnected neurons, including both excitatory principal cells and a diverse assortment of regulatory interneurons. As neurons communicate through electrochemical mechanisms, characterizing the electrical signatures of both control and experimentally perturbed neurons provides great insight into the properties of the fundamental components that compose brain circuits.
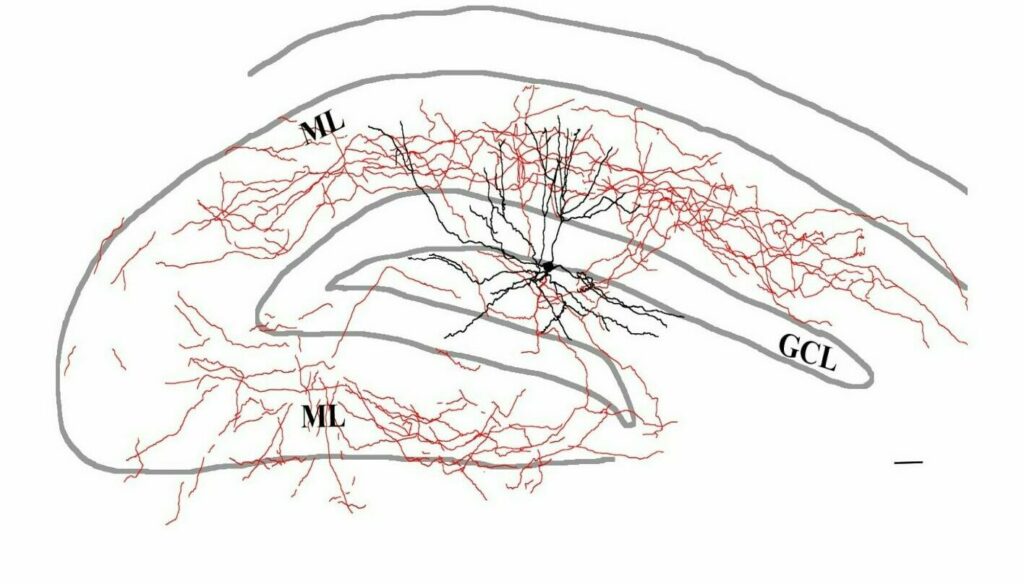
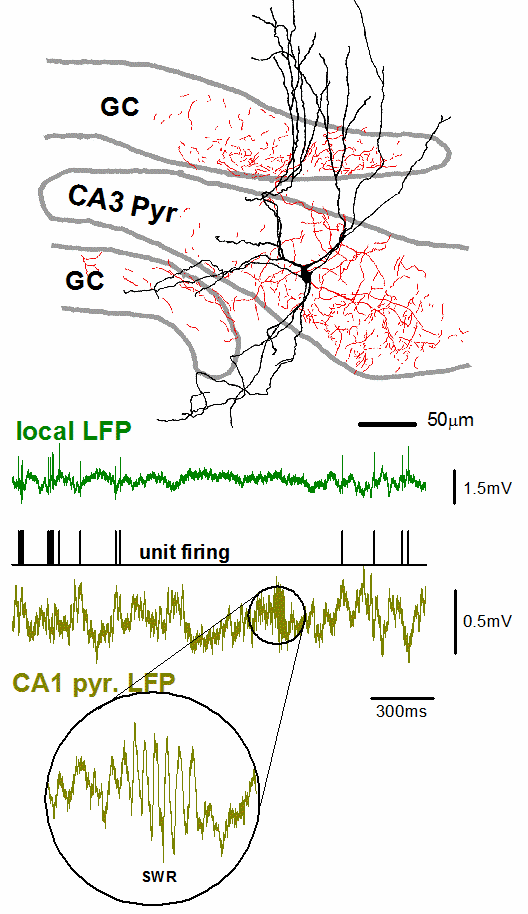
With whole cell electrophysiology, we use finely pointed glass electrodes to precisely record the electrical signal from within individual neurons. Thus, we can examine both the functional electrical properties intrinsic to that particular cell and monitor the synaptic inputs that neuron receives. While recordings are often made from the neuronal cell body, the Soltesz lab has experience recording specifically from smaller structures, including hippocampal dendrites or even axonal terminals.
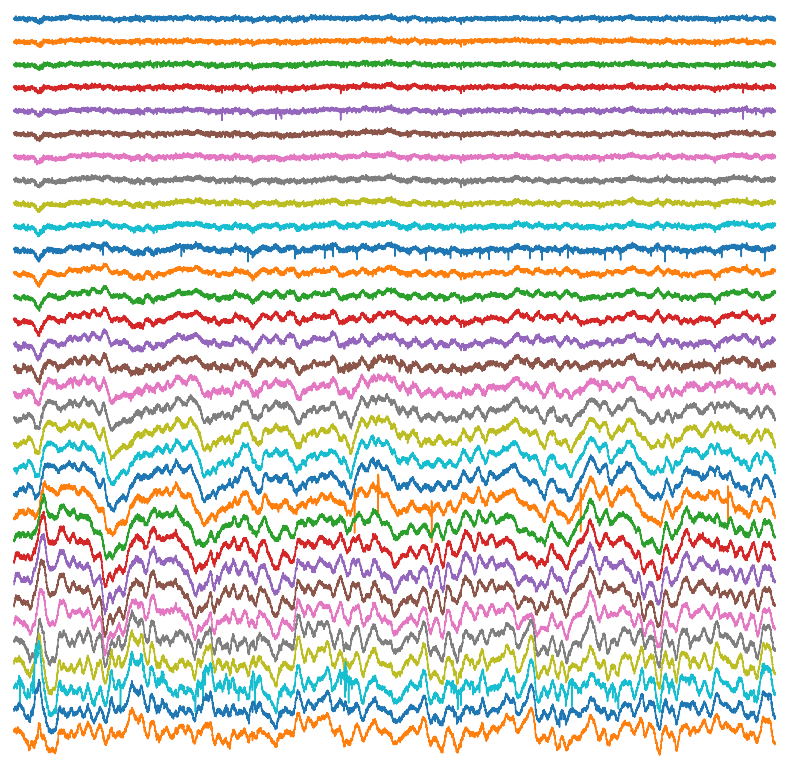
We also utilize several variants on the whole cell electrophysiology approach to interrogate other features of neuronal networks. We can apply compounds to manipulate neuronal activity either through bath application onto the entire brain slice, or through more targeted application via a microinjection system. Through paired patch-clamp recordings, we explore how two simultaneously recorded neurons (whether of the same or different cell types) are interconnected and how they influence the electrical properties of each other. We also employ optogenetic techniques to alter the activity of genetically targeted neuronal populations in response to light stimulation, both to examine their downstream connectivity and how they influence the electrical properties of other neurons. Furthermore, we have the capacity to combine whole cell electrophysiology with simultaneous high-speed imaging of optical probes (e.g. calcium and voltage indicators), enabling us to incorporate the high single cell precision of electrophysiology with broader population level readouts of activity within a brain slice.
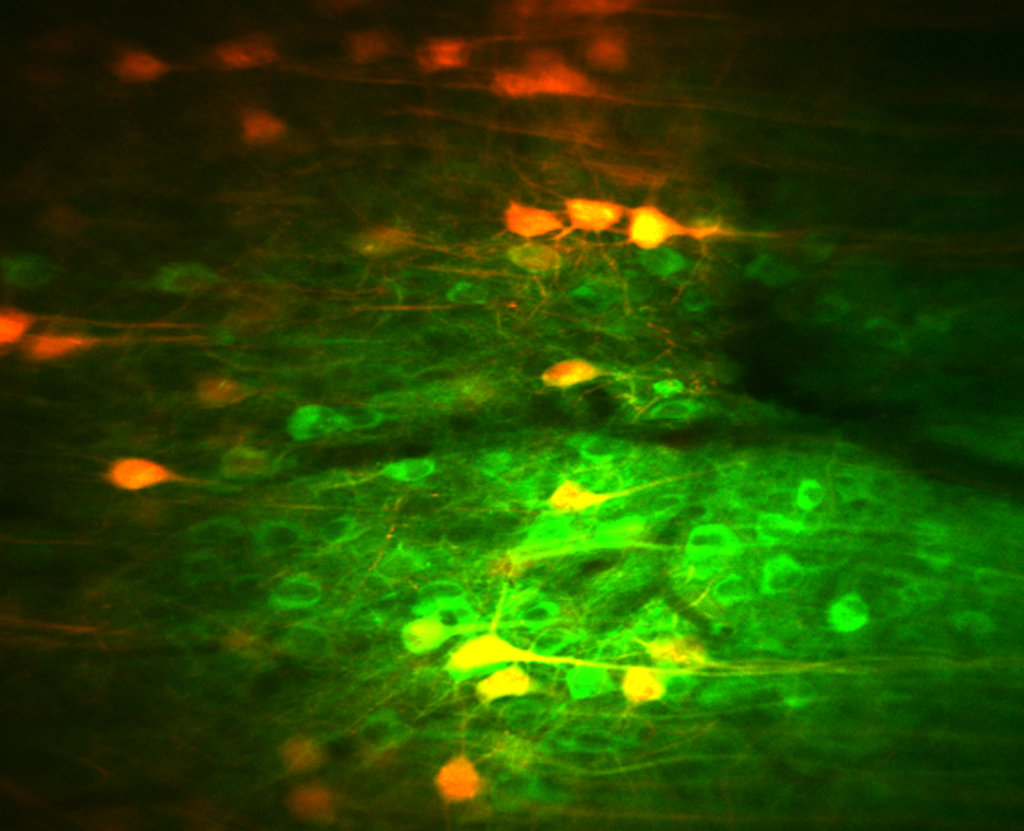
Neurons recorded with whole cell electrophysiology can be filled with intracellular tracers, allowing enabling later morphological reconstruction of the cell. Subsequently, the labelled cell can be further stained for neurochemical markers with immunohistochemistry.
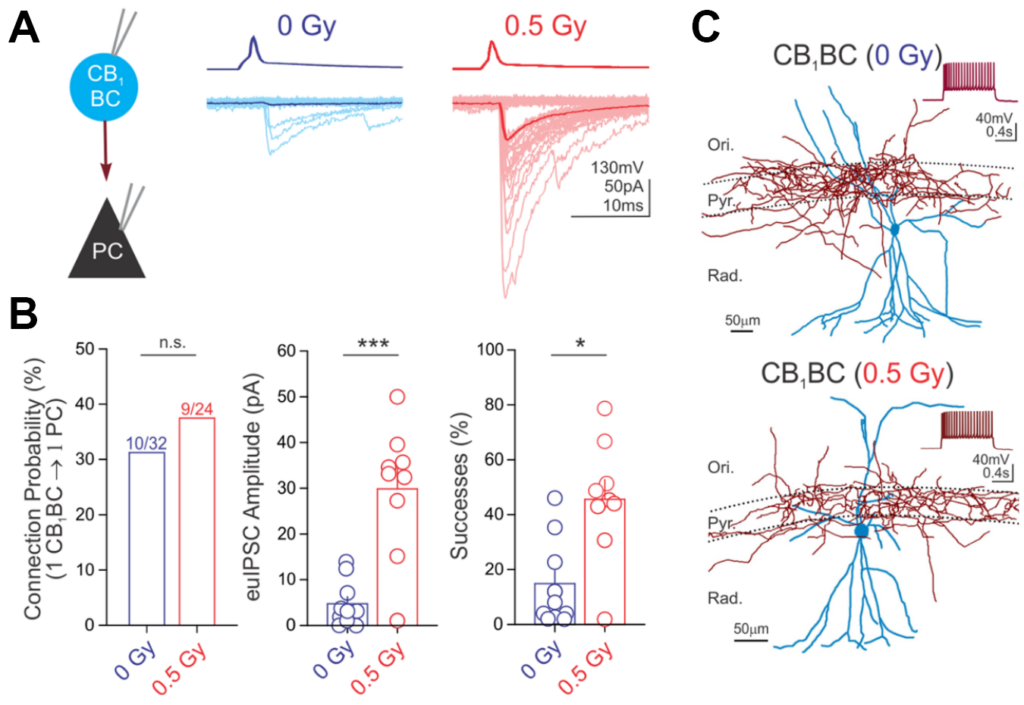
Below is an example of data from paired recordings of hippocampal interneuron (CB1BC) signaling to pyramidal cells (PC), where CB1BC stimulation evoked inhibitory currents in the PC (panel A). The impact of a manipulation on CB1BC to PC signaling, in this case following proton irradiation (0.5 cGy) was then quantified (panel B). The morphology of the recorded neurons was then reconstructed (panel C). For further information, see Lee S-H et al., 2017, Brain Structure and Function.